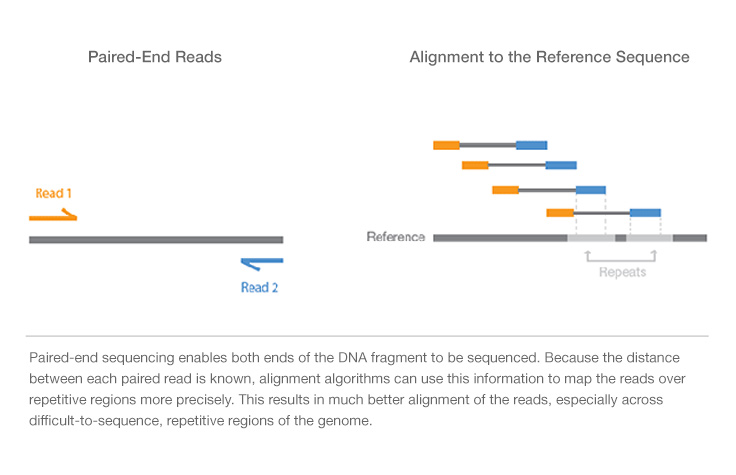
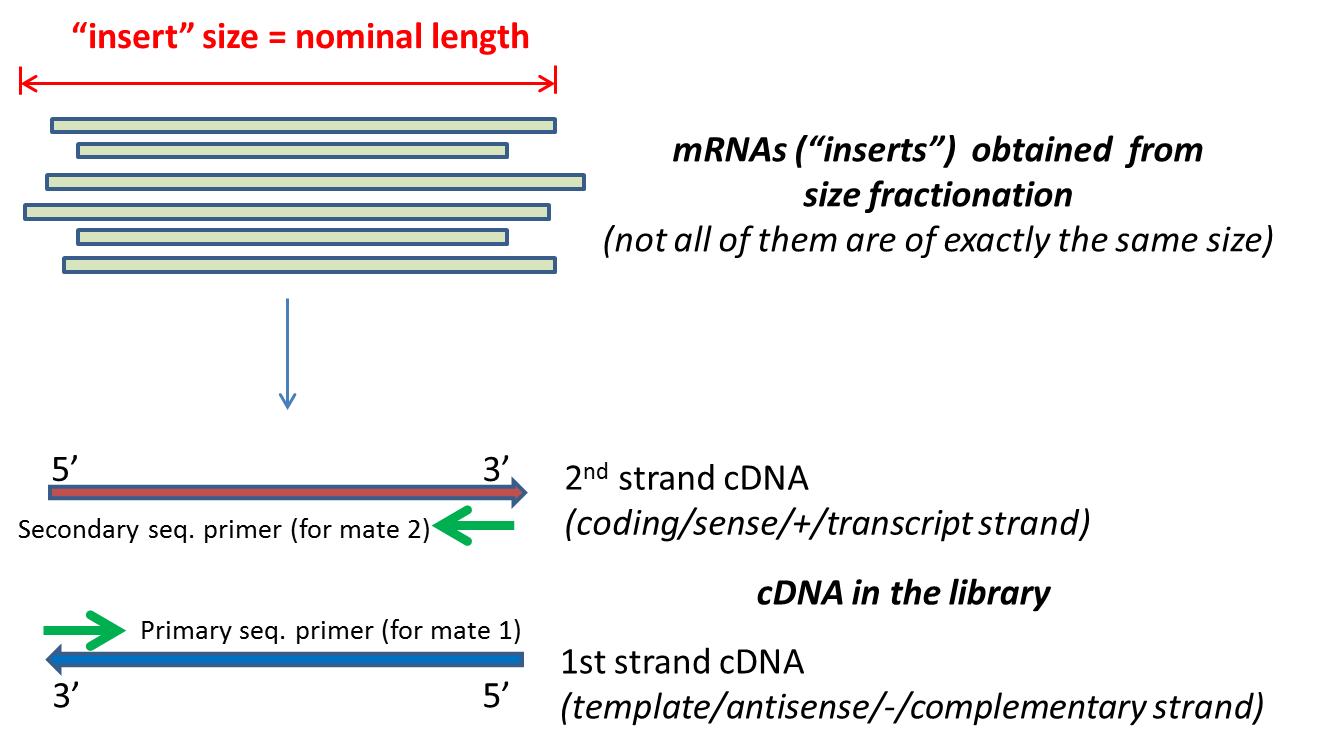
paired end sequencing insert size
Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size. The insert size on classic paired-end is smaller about 500bp while the insert size of mate-pair is much longer several Kb which allows to join the contiguous between them especially is it.
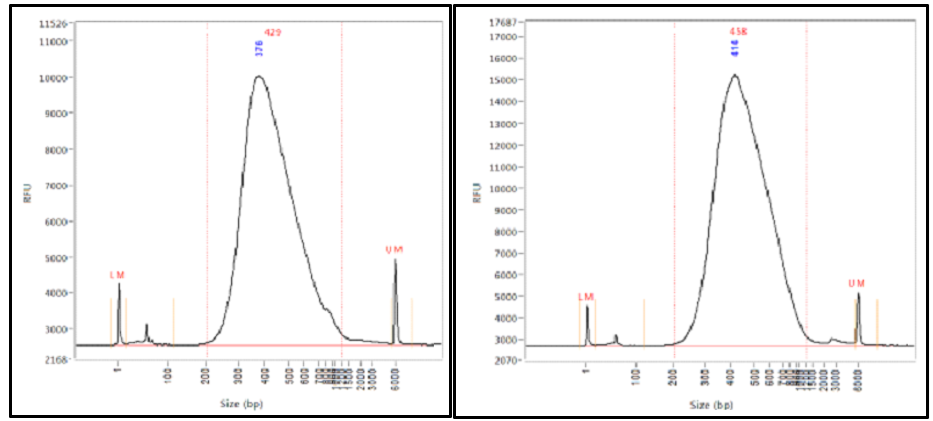
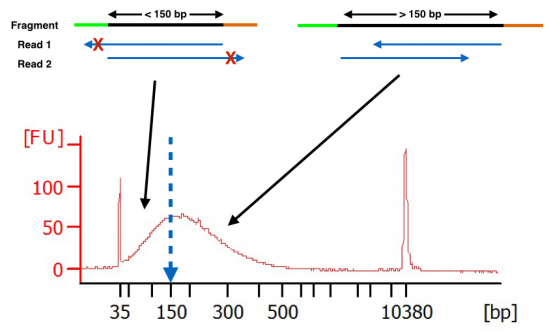
How Short Inserts Affect Sequencing Performance
However the average size of human coding exons is only 160bp.

. Adapter sequences will add an additional 130 to. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size. For example a 300-cycle kit can be used for a 1 300 bp single-read run or a 2 150 bp paired-end run.
If processing a small file set the minimum percentage option M to 05 otherwise an error may occur. As we know by now and the insert size limitation becomes an issue when you want paired-end reads with a higher inner distance with fragments longer than 800 bp. Including flanking intronic regions of diagnostic significance eg due to splice variants of about 30bp at either side the average target of interest is about 220 bp long.
The fragment size which you need to select for during a gel purification for example would be the insert size length of both adapters around 120 bp extra for both Illumina. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece.
Broad Range of Applications. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. In example 2 the inferred insert size is.
Here we modify the Illumina RNA ligation protocol to allow SS-PE sequencing by using a custom pre-adenylated 3 adaptor. 2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and maximum insert-length as 500 or more. Here we modify the Illumina RNA ligation protocol to allow SS-PE sequencing by using a custom pre-adenylated 3â adaptor.
Please see InsertSizeMetrics for detailed explanations of each metric. Get 1 month free of our Silver Membership including 2 additional DNA reports. Ad Access more DNA discoveries than has ever before been possible with Sequencing.
You can use sequencing reagents to generate single continuous reads or for paired-end sequencing in both directions. The fragment size which you need to select for during a gel purification for example would be the insert size length of both adapters around 120 bp extra for both Illumina adapters. BWA insert size calculation in paired end sequencing 1.
Length of the entire amplicon insert. Example 1 - inferred insert size is rightmost coordinate read length - leftmost coordinate. For instance for a PE150 sequencing run the insert size should be above 300bp.
Leftmost coordinate read length - rightmost coordinate. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. 31 rows java -jar picardjar CollectInsertSizeMetrics.
Ranges from 2 150 to 2 300 bp. For instance for a PE150 sequencing run the insert size should be above 300bp. As an alternative there is a preparation method called Mate pair sequencing which enables you to use fragments up to 5 kba.

Interpreting Color By Insert Size Integrative Genomics Viewer
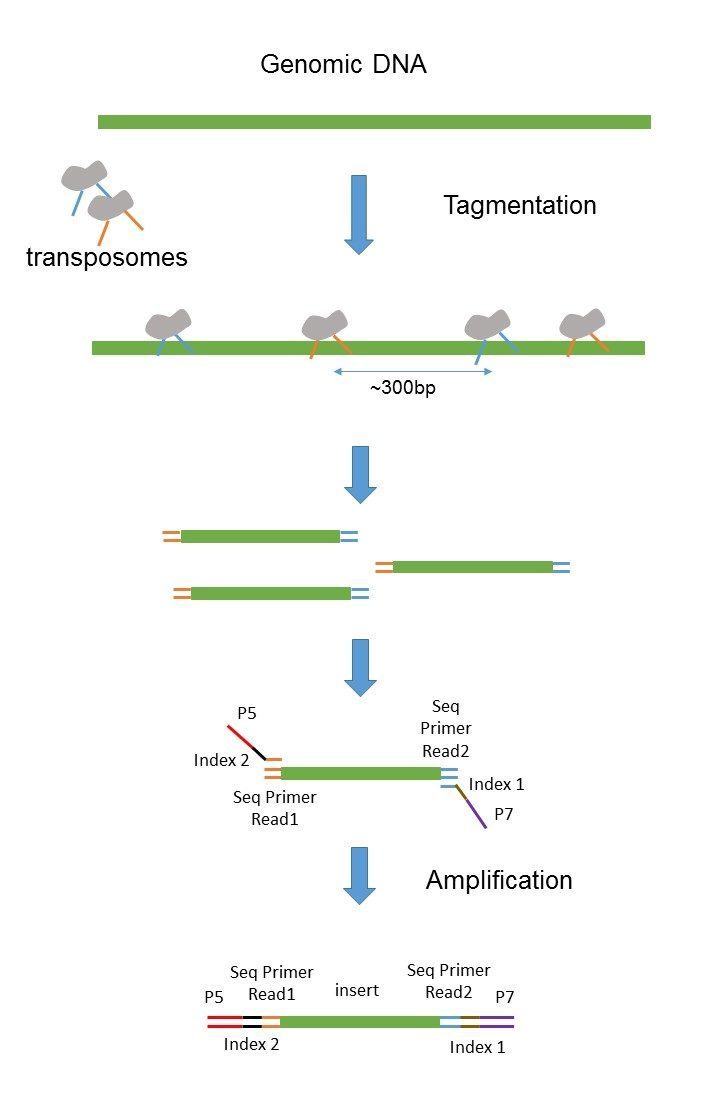
Paired End Sequencing France Genomique

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

What Are Paired End Reads The Sequencing Center
The Insert Size In Paired End Data Seqanswers

Library Insert Size Libraries Were Constructed Using The Standard A Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer
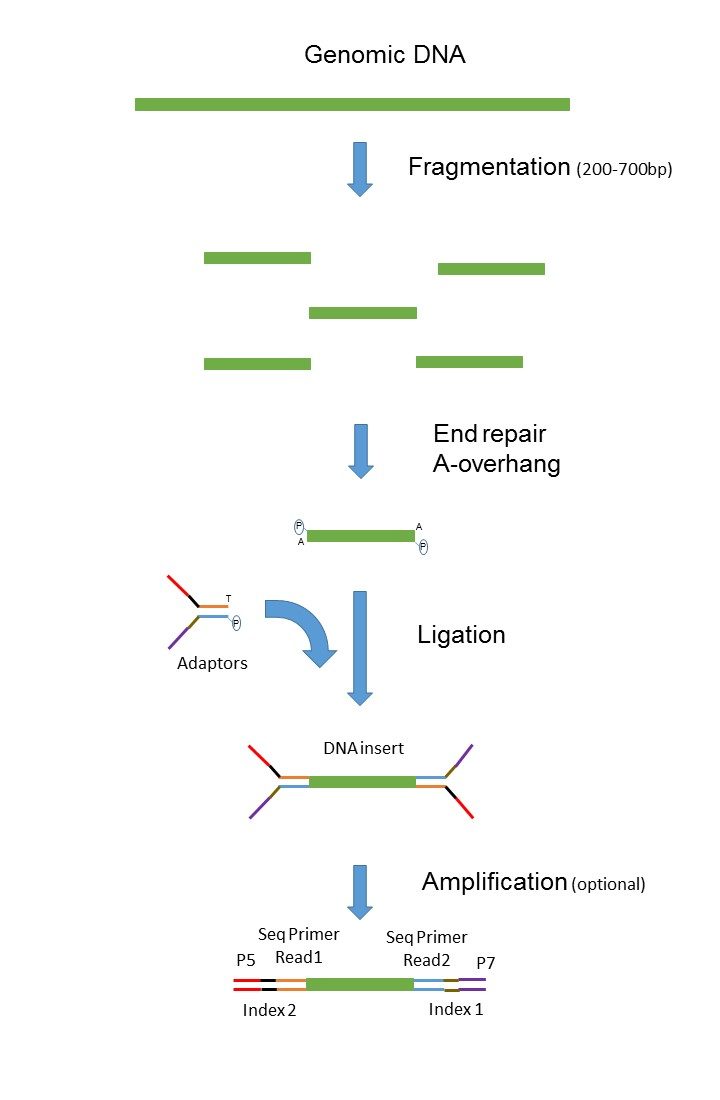
Paired End Sequencing France Genomique

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram

What Is Mate Pair Sequencing For

What Are Paired End Reads The Sequencing Center
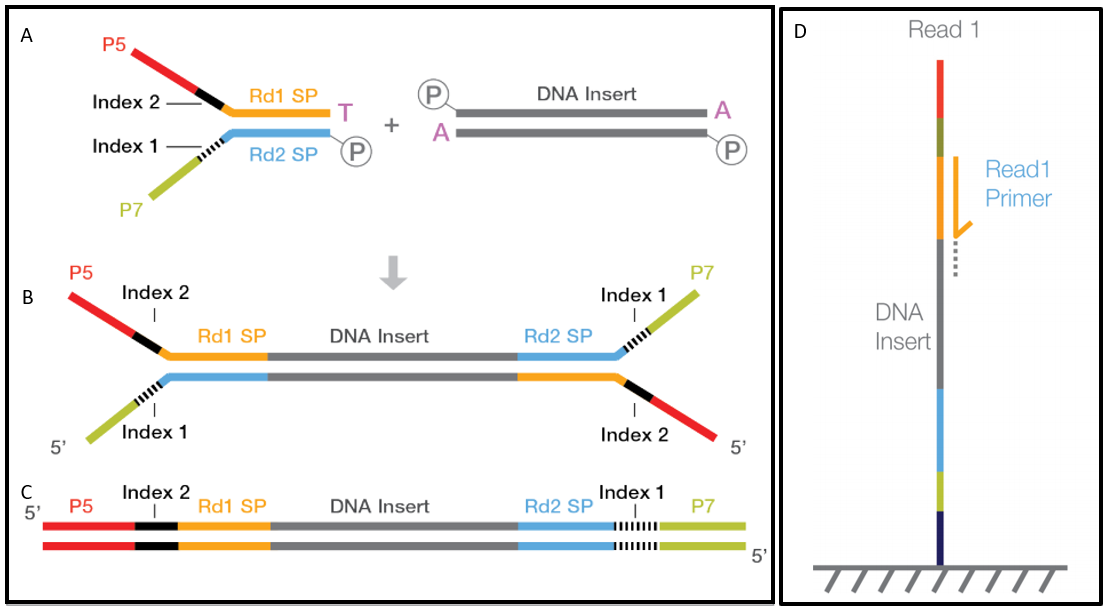
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

What Is Mate Pair Sequencing For
What Is Paired End 150 Omega Bioservices
How Short Inserts Affect Sequencing Performance

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram